Guix-HPC Activity Report, 2023

This document is also available as PDF (printable booklet)
We are pleased to publish the sixth Guix-HPC annual report. Launched in 2017, Guix-HPC is a collaborative effort to bring reproducible software deployment to scientific workflows and high-performance computing (HPC). Guix-HPC builds upon the GNU Guix software deployment tool to empower HPC practitioners and scientists who need reliability, flexibility, and reproducibility; it aims to support Open Science and reproducible research.
Guix-HPC started as a joint software development project involving three research institutes: Inria, the Max Delbrück Center for Molecular Medicine (MDC), and the Utrecht Bioinformatics Center (UBC). GNU Guix for HPC and reproducible research has since received contributions from many individuals and organizations, including CNRS, Université Paris Cité, the University of Tennessee Health Science Center (UTHSC), Cornell University, and AMD. HPC remains a conservative domain but over the years, we have reached out to many organizations and people who share our goal of improving upon the status quo when it comes to software deployment.
This report highlights key achievements of Guix-HPC between our previous report a year ago and today, February 2024. This year was marked by exciting developments for HPC and reproducible workflows: the organization of a three-day workshop in November on this very topic where 120 researchers and HPC practitioners met, the expansion of the package collection available to Guix users—including significant contributions by AMD and new channels giving access to all of Bioconductor, along with more ground work to meet the needs of HPC and reproducible research.
Outline
Guix-HPC aims to tackle the following high-level objectives:
- Reproducible scientific workflows. Improve the GNU Guix tool set to better support reproducible scientific workflows and to simplify sharing and publication of software environments.
- Cluster usage. Streamlining Guix deployment on HPC clusters, and providing interoperability with clusters not running Guix.
- Outreach & user support. Reaching out to the HPC and scientific research communities and organizing training sessions.
The following sections detail work that has been carried out in each of these areas.
Reproducible Scientific Workflows
Supporting reproducible research workflows is a major goal for Guix-HPC. This section looks at progress made on packaging and tooling.
Packages
The package collection available from Guix keeps growing: as of this writing, Guix itself provides more than 29,000 packages, all free software, making it the fifth largest free software distribution. With the addition of scientific computing channels, users have access to more than 52,000 packages!
We updated the hpcguix-web package browser and its Guix-HPC instance to make it easier to search these channels, to navigate them, and to get set up using them. The channels page lists channels commonly used by the scientific community. A noteworthy example is Guix-Science, now home to hundreds of packages. Most of these channels are under continuous integration, with pre-built binaries being published from build farms such as that hosted by Inria.

Expanding on the introduction of the
guix-cran channel last
year, we are happy to announce the new
guix-bioc channel.
This new channel makes most of the entire
Bioconductor collection of R packages
available as Guix packages. Substitutes are provided by the build
farm at guix.bordeaux.inria.fr to speed up installation times. The
channel augments the collection of R packages provided by the
Guix default channel and the guix-cran channel. Creating and
updating guix-bioc is fully automated and happens without any human
intervention. The channel itself is always in a usable state, because
updates are tested with guix pull before committing and pushing
them. The same limitations of the guix-cran channel with regard to
potential build failures due to undeclared build or runtime
dependencies also apply to this channel. Improvements to the CRAN
importer in Guix, however, have allowed us to reduce the failure rate
and raise the quality of both channels.
These two automated channels grow the number of R packages available in reproducible Guix environments by 21,635 to a total of 24,187. Unlike other efforts that aim to provide binaries of R packages, the collection of R packages in Guix fully captures all dependencies, including those that would otherwise be considered “system dependencies”, insulating Guix environments from system-level changes over time. The increasing coverage of package sources archived by Software Heritage puts Guix in a unique position as a solid foundation for reliable long-term reproducible research with R.
A major highlight this year is the 100+ packages contributed by AMD for its ROCm and HIP toolchain for GPUs. Those include 5 versions of the entire HIP/ROCm toolchain, all the way down to LLVM and including support in communication libraries ucx and Open MPI. Anyone who has tried to package or to build this will understand that this is a major contribution: the software stack is complex, requiring careful assembly of the right versions or variants of each component.
Those packages are a boost to the supercomputer users. We have been
able to use them to run HIP/ROCm benchmarks on the French national
supercomputer
Adastra,
which features AMD Instinct MI250X GPUs, leveraging guix pack to ship
the code. We expect this joint effort with AMD to continue so we can
deliver other parts of the stack—e.g., rocBLAS, rocFFT, and related math
libraries—and to enable ROCm support in other packages such as PyTorch and Tensorflow.
For those systems where the HIP/ROCm stack cannot be used, the Guix Science Nonfree channel provides various versions of CUDA and cuDNN. This channel now also provides CUDA-enabled variants of packages from the Guix Science channel that only support CPU-based inference. Of note is the addition of both the CPU- and CUDA-enabled variants of JAX, the machine learning framework for accelerated linear algebra and automated differentiation of numerical functions. Recent versions of Tensorflow 2 and related Tensorflow libraries are now also available, thanks to the addition of a Bazel build system abstraction in the Guix Science channel.
Other notable additions to the Guix-HPC channel include the plethora of dependencies needed to build GEOS, a geophysical simulation framework, and medInria, a medical image processing and visualization package, both contributed by Inria engineers.
Guix Packager, a Packaging Assistant
Defining packages for Guix is not all that hard but, as always, it is much harder the first time you do it, especially when starting from a blank page or not being familiar with the programming environment of Guix. Guix Packager is a new web user interface to get you started.
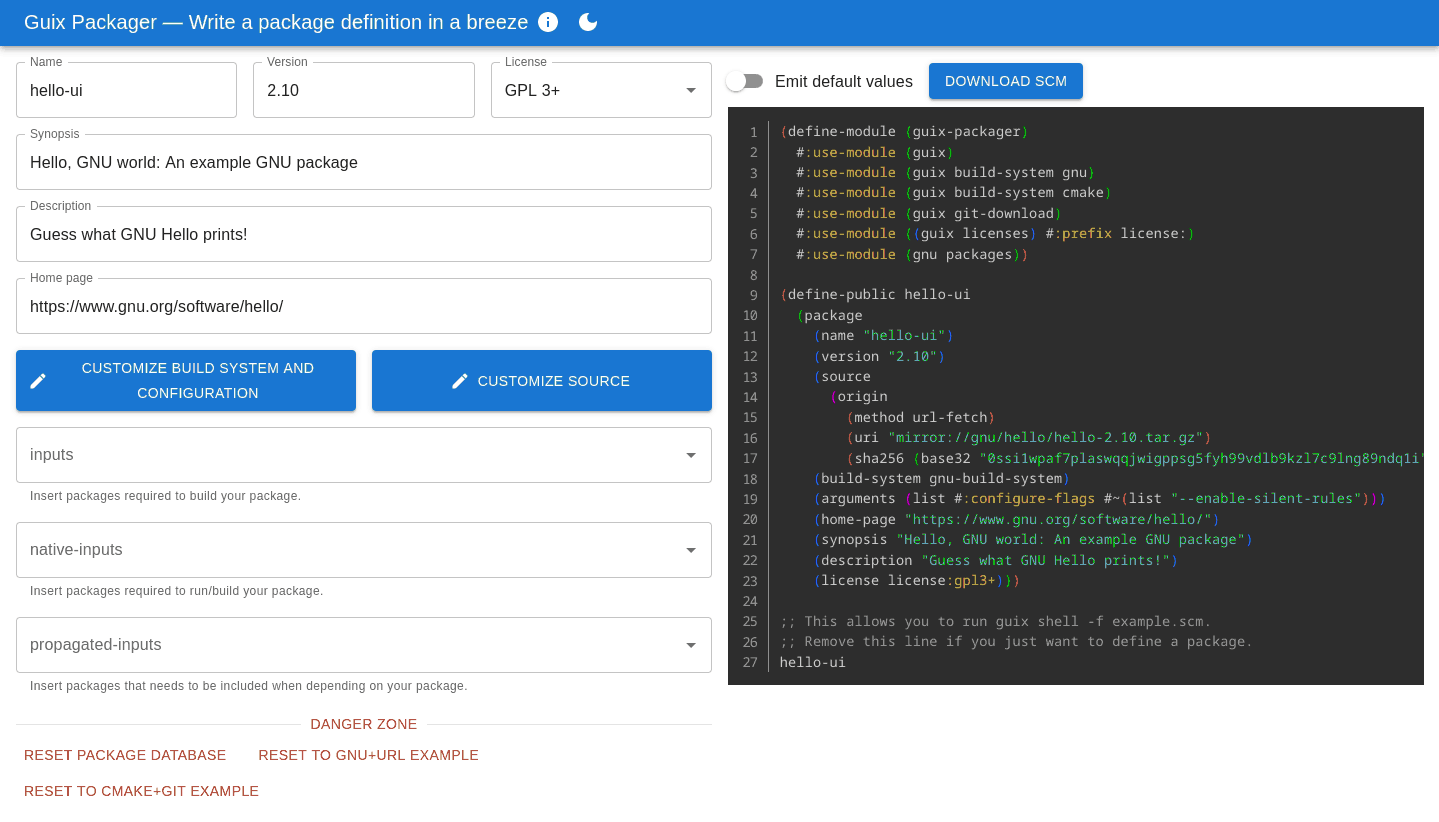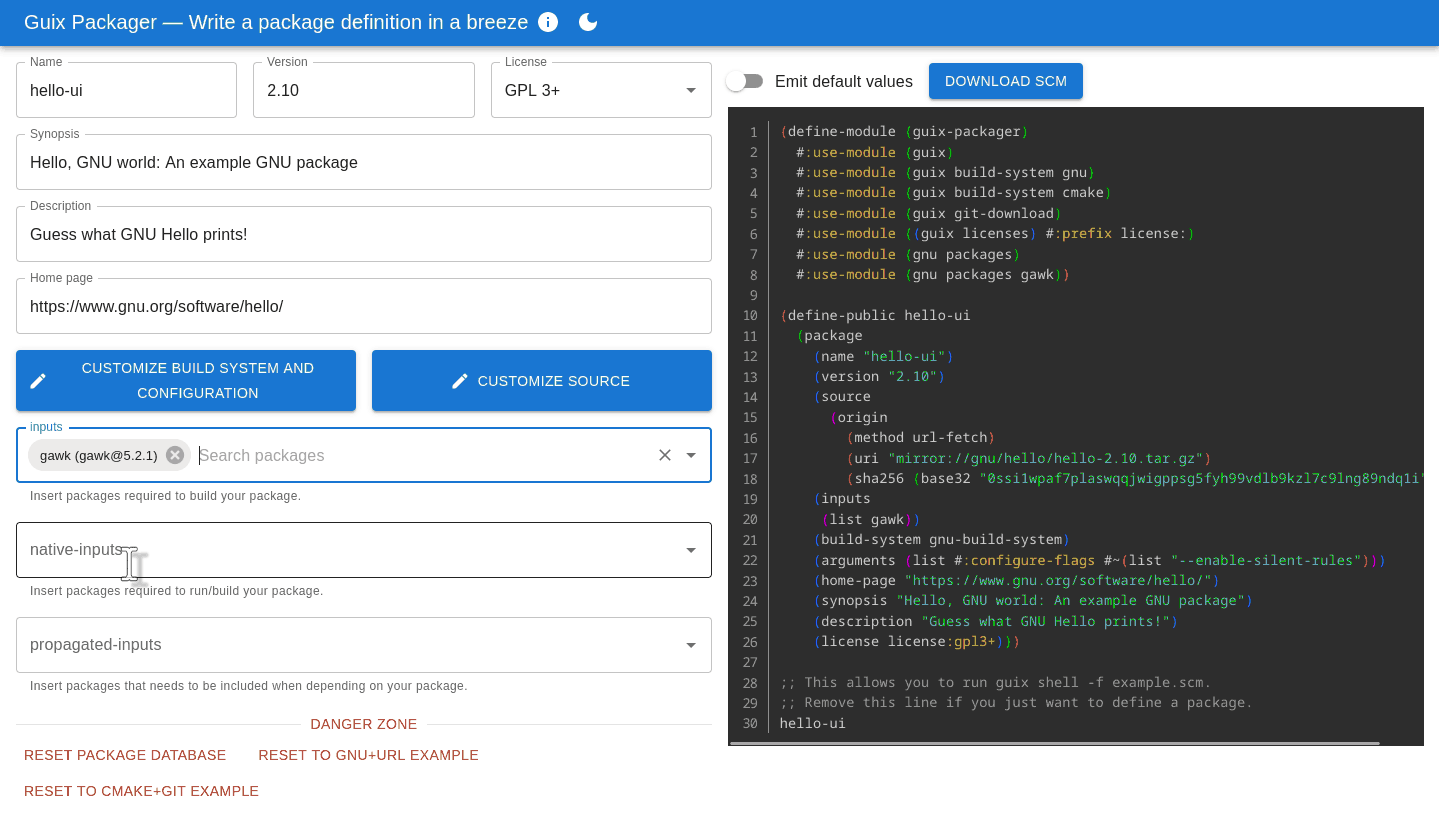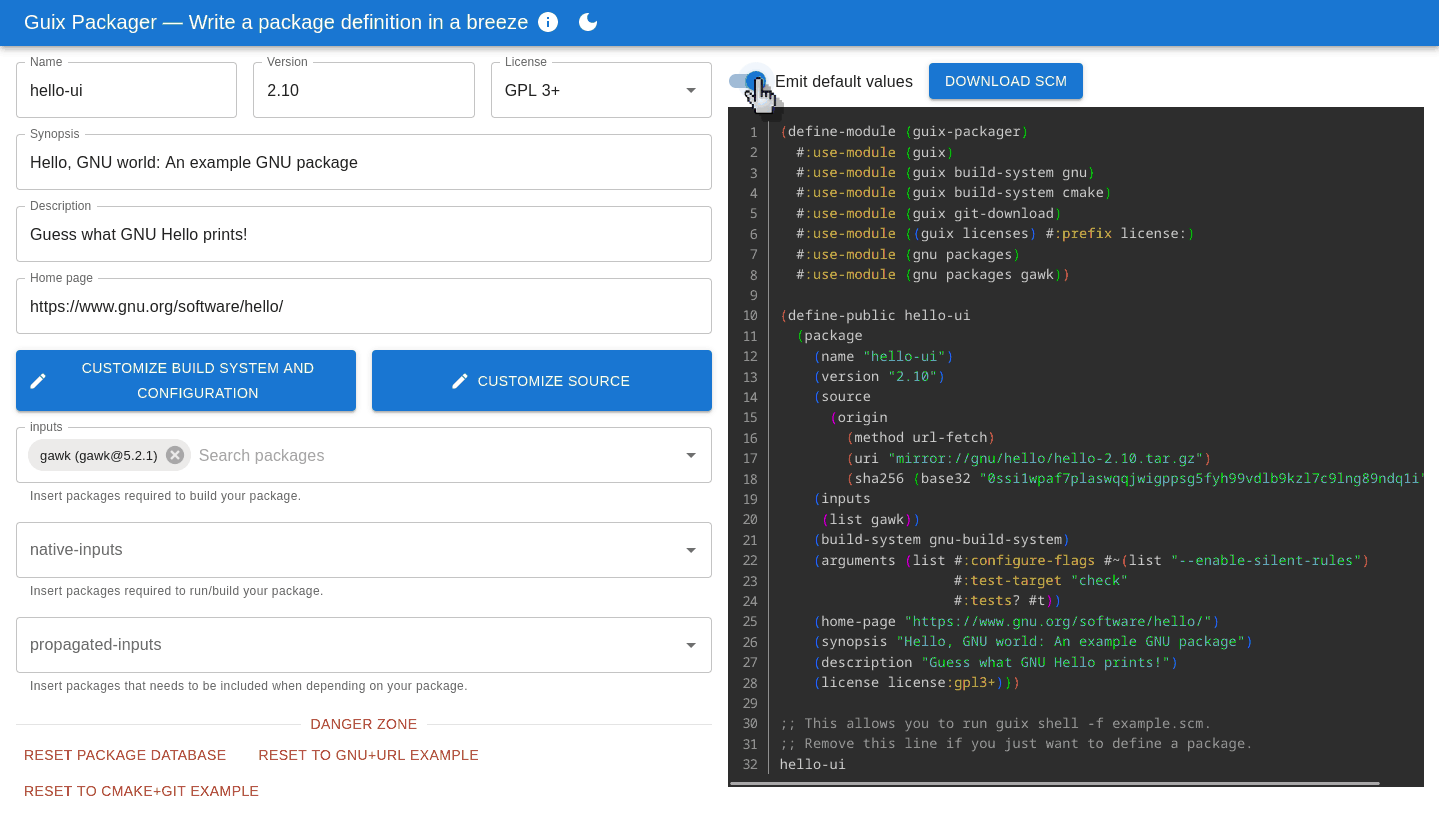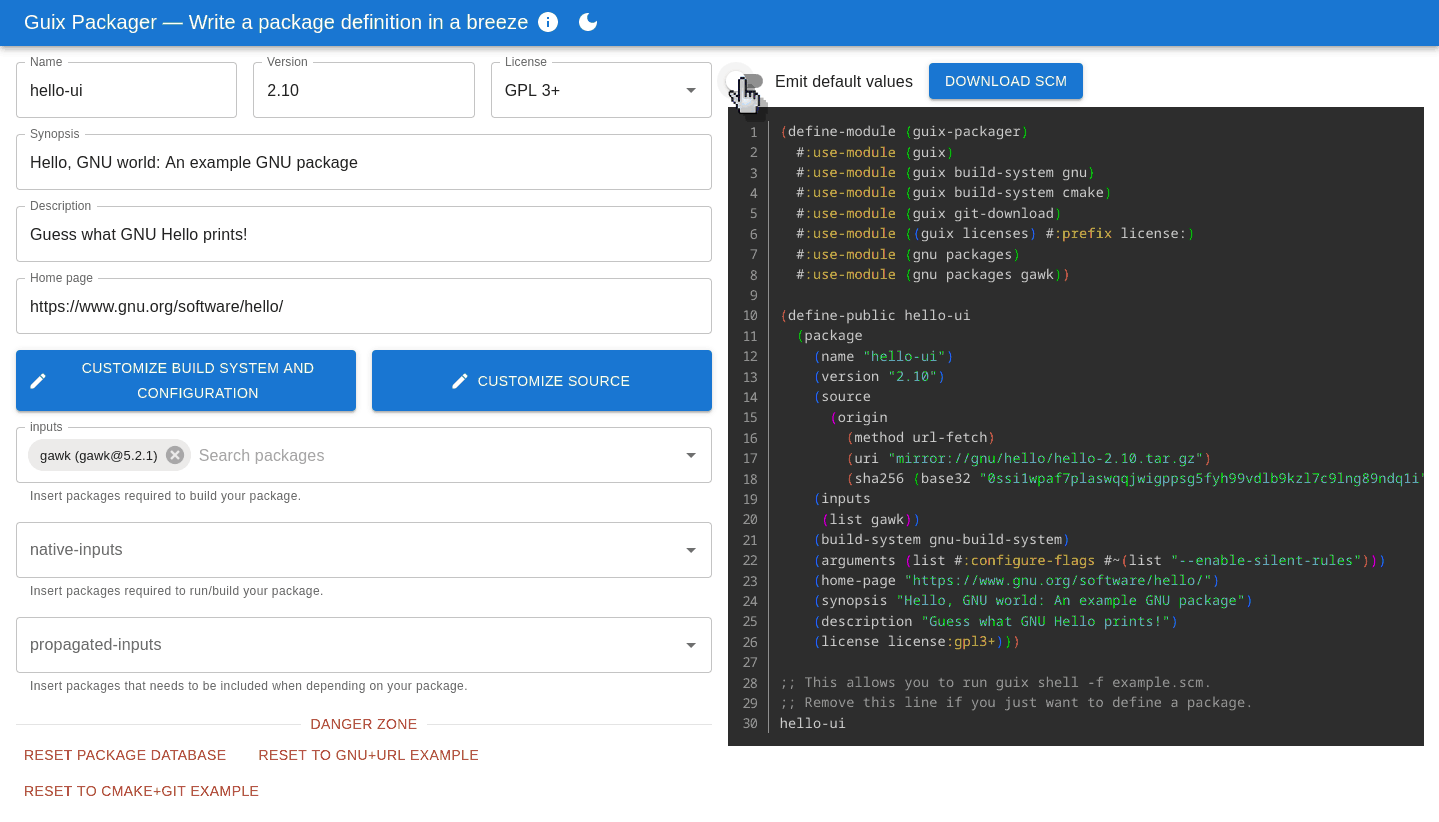
The interface aims to be intuitive: fill in forms on the left and it produces a correct, ready-to-use package definition on the right. Importantly, it helps avoid pitfalls that trip up many newcomers: adding an input adds the right variable name and modules, turning tests on and off or adding configure flags can be achieved without prior knowledge of the likes of keyword arguments and G-expressions.
While the tool's feature set provides a great starting point, there are still a few things that may be worth implementing. For instance, only the GNU and CMake build systems are supported so far; it would make sense to include a few others (Python-related ones might be good candidates).
Ultimately, Guix Packager does not intend to provide a full package definition
editor, but rather a simple entry point for people looking into starting to
write packages definitions.
It complements a set of steps we've taken over time to make packaging in Guix
approachable. Indeed, while package definitions are
actually code written in the Scheme language, the package “language”
was designed from the get-go to be
fully declarative—think JSON with parentheses instead of curly braces and
semicolons.
Nesting Containerized Environments
The guix shell --container
(or guix shell -C) command lets users create isolated software
environments—containers— providing nothing but the packages specified
on the command line. This has proved to be a great way to ensure the
run-time environment of one’s software is fully controlled, free from
interference from the rest of the system.
Recently though, a new use case came up, calling for support of nested containers. As Konrad Hinsen explained, the need for nested containers arises, for example, when dealing with workflow execution engines such as Snakemake and CWL: users may be willing to use Guix to deploy both the engine itself and the software environment of the tasks the engine spawns.
This is now possible thanks to the new --nesting or -W option, to be
used in conjunction with --container or -C. This option lets users
create nested containerized environments as in this example:
guix shell --container --nesting coreutils -- \
guix shell --container pythonThe “outer” shell creates a container that contains nothing but
coreutils—the package that provides ls, cp, and other core
utilities; the “inner” shell creates a new container that contains
nothing but Python. For a Snakemake workflow, one would run:
guix shell --container --nesting snakemake -- \
snakemake …… which in turn allows the individual tasks of the workflow to run guix shell as well.
Concise Common Workflow Language
The Concise Common Workflow Language
(ccwl) is a concise syntax to
express Common Workflow Language (CWL) workflows. It is implemented as an
EDSL (Embedded Domain Specific Language) in Guile Scheme. Unlike workflow
languages such as the Guix Workflow Language (GWL), ccwl is agnostic to
deployment. It does not use Guix internally to deploy applications. It
merely picks up applications from PATH and thus interoperates well with Guix
and any other package managers of the user's choice. ccwl also compiles to
CWL and thus reuses all tooling built to run CWL workflows. Workflows written
in ccwl may be freely reused by CWL users without impediment, thus ensuring
smooth collaboration between ccwl and CWL users.
ccwl 0.3.0 was released in January 2024. ccwl 0.3.0 comes with significantly better compiler error messages to detect errors early and provide helpful error messages to users. ccwl 0.3.0 also adds new constructs to express scattering workflow steps and other more complex workflows.
Ensuring Source Code Availability
Our joint effort with Software Heritage (SWH) has made major strides this year on the two main fronts: increasing archive coverage, and improving source code recovery capabilities. The two are closely related but involve different work; together, they contribute to making Guix a tool of choice for reproducible research workflows.

Timothy Sample has been leading the archival effort and closely monitoring it. His latest Preservation of Guix Report, published in January 2024, reveals that 94% of the package source code referred to by Guix at that time is archived in SWH. That number has been steadily increasing since we started this effort in 2019. Archival coverage for the entire 2019–2024 period is 85%. Having identified the missing bits, the SWH team is now retroactively ingesting package source code of historical Guix revisions.
Guix’s ability to recover source code from SWH has improved in part
thanks to the newly-added support for bzip2-compressed archives in
Disarchive, the tool
designed to allow Guix to recover exact copies of source code tarballs
such as .tar.gz and tar.bz2 files.
A longstanding issue for automatic recovery from SWH is a mismatch
between the cryptographic hashes used in Guix and in SWH to refer to
content—a problem identified early
on.
This has been addressed by a recent SWH feature deployed in
January 2024: SWH now computes and exposes nar SHA256 hashes for
directories—the very hashes used in Guix package definitions. Those
hashes are added as an extension of the SWH data model called external
identifiers or ExtIDs; the HTTP interface lets us obtain the SWHID
corresponding to a nar-sha256 ExtID, which is exactly what was
necessary to ensure content-addressed access in all cases.
Consequently, the fallback code in Guix was changed to use that method.
This will allow Guix to recover source code for version control systems
(VCS) other than Git, which was previously not possible.
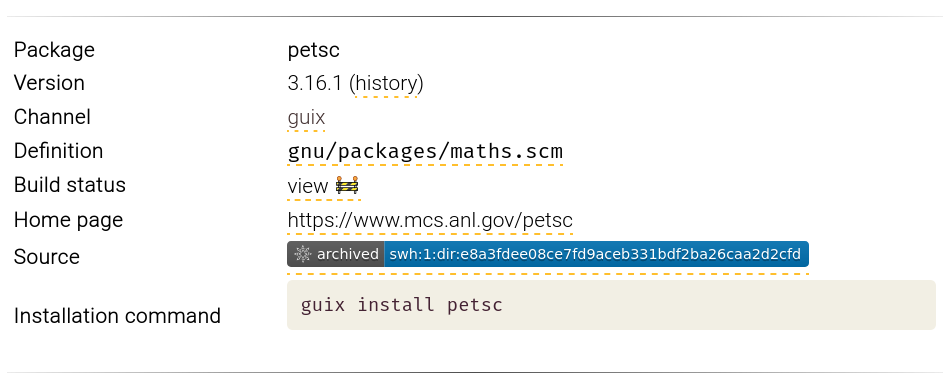
To make SWH archival more tangible to users and packagers, we modified
the hpcguix-web package browser, visible on the Guix-HPC web
site, to include a source code archival
badge on every package page. The badge, served by SWH, is currently
shown both for packages whose source code is fetched from a Git
repository, and for packages whose source code is fetched from a
tarball. The information is comparable to that checked by the guix lint -c archival command.
Reproducible Research in Practice
In February 2023, Marek Felšöci defended his PhD thesis entitled Fast solvers for high-frequency aeroacoustics. The thesis was part of a collaboration between Inria and Airbus and deals with direct methods for solving coupled sparse/dense linear systems. Chapter 8 of the manuscript explains the strategy that was used to achieve reproducible and verifiable results and how Guix, Software Heritage, and other tools support it. It is another testimonial showing how reproducible computational workflows can be achieved, even in a demanding HPC context.
In a talk entitled Everyone Can Learn How to Guix, medical doctor Nicolas Vallet defended a similar thesis: tools such as Guix can support reproducible research workflows and be viewed as key enablers even in scientific domains one might think of as detached from software deployment considerations.
NumPEx is the French national program for exascale HPC, launched in mid-2023 with a 41 M€ budget for 6 years. Its Development and Integration project aims to ensure the dozens of HPC libraries and applications developed by French researchers can easily be deployed on national and European clusters, with high quality assurance levels. Guix is one of the deployment tools used to achieve those goals and well poised to do so. The project has just recruited two engineers to help with packaging, continuous integration, and training in this context.
We hope this will not only help create synergies with the broader Guix
community, but also contribute to increasing awareness about
reproducible deployment in HPC circles. Meanwhile, conducting
reproducible research on supercomputers that lack Guix is already possible:
by creating an image with guix pack, deploying it on
the supercomputer, and setting up the host environment properly.
Experiments have shown that it
does not lead to any significant performance difference compared to
the same code and software stack deployed natively. The motivation,
technical details, and performance study were presented in a talk
entitled Reconciling high-performance computing with the use of
third-party
libraries?
Another aspect related to reproducibile HPC research and development is the environment used to write code, document it, post-process data, produce scientific reports. Offering researchers and developers a way to share the exact same working environment is one way to facilitate collaboration. The Elementary Emacs configuration coupled with Guix (ElementaryX) project is an attempt towards such an elementary yet reproducible environment.
Cluster Usage and Deployment
The sections below highlight the experience of cluster administration teams and report on tooling developed around Guix for users and administrators on HPC clusters.
Usage at the German Aerospace Center
The Institute of Networked Energy Systems of the German Aerospace Center (DLR) has set up a Guix installation in its HPC system and transitioned several workflows to Guix, which are related to remote sensing and solar surface radiation and feed data into the European Copernicus Atmosphere Monitoring Service CAMS. Similar to containers, Guix software stacks are almost independent of the host system. However, container support in HPC systems is limited and still evolving. Guix relocation options offer more flexibility and the software stack has been successfully deployed in HPC clusters available to the DLR (like CARA, CARO and terrabyte), thereby enabling easy scaling of the radiation services.
Guix System Cluster at GLiCID
GLiCID is the HPC center for research in the French region Pays de la Loire, resulting from the merger of pre-existing HPC centers in the region.
The installation of new machines in June 2023 has led to the launch of a new common system infrastructure—identity management, SLURM services, databases, etc.—mostly independent from the solutions provided by the manufacturers. Installed on two remote data centers, the infrastructure needs to be highly available, and its deployment can be complex. The team wanted to guarantee simple, predictable redeployment of the infrastructure in the event of problems.
Guix, already offered to all cluster users, has a proven track record of reproducibility, a desirable feature not just for scientific software but also for the infrastructure itself. That is why the team embarked on an effort to build its infrastructure with Guix System, which led to the development of Guix System services for HPC—for OpenLDAP, SLURM, and more. They reported on the impact of these choices at the Workshop in Montpellier, and are currently making progress to reach a 100% Guixified infrastructure.
Pangenome Genetics Research Cluster at UTHSC
At UTHSC, Memphis (USA), we are running a 16-node large-memory Octopus HPC cluster (438 real CPU cores) dedicated to pangenome and genetics research. In 2023, the cluster effectively doubled in size with 192 4 GHz CPU cores, 144,000 GPU cores, and SSDs added. The storage adding up to a 200 TB Lizardfs fiber-optic connected distributed network storage.
Notable about this HPC cluster is that it is administered by the users themselves. Thanks to Guix, we install, run and manage the cluster as researchers—and roll back in case of a mistake. UTHSC IT manages the infrastructure—i.e., physical placement, electricity, routers and firewalls—but beyond that there are no demands on IT. Thanks to out-of-band access, we can completely (re)install machines remotely. Octopus runs Guix on top of a minimal Debian install and we are experimenting with pure Guix virtual machines and nodes that can be run on demand. Almost all deployed software has been packaged in Guix and can be installed on the head-node by regular users on the cluster without root access. This same software is shared through NFS on the nodes. See the guix-bioinformatics channel for all deployment configuration.
At FOSDEM 2023, Arun Isaac presented Tissue, our minimalist Git+plain text issue tracker that allows us to move away from GitHub source code hosting, continuous integration (CI), and issue trackers. We have also started to use Guix with the Concise Common Workflow Language (CCWL) for reproducible pangenome workflows (see above) on our Octopus HPC.
Supporting RISC-V
RISC-V is making inroads with HPC, e.g. in Barcelona and with the new Barcelona Supercomputing Center Sargantana chip.
Christopher Batten (Cornell) and Michael Taylor (University of Washington) are in charge of creating the NSF-funded RISC-V supercomputer with 2,000 cores per node and 16 nodes in a rack (NSF PPoSS grant 2118709), targeting Guix driven pangenomic workloads by Erik Garrison, Arun Isaac, Andrea Guarracino, and Pjotr Prins.
The supercomputer will incorporate Guix and the GNU Mes bootstrap, with input from Arun Isaac, Efraim Flashner and others.
NLNet funds RISC-V support for the Guix riscv64 target from Efraim Flashner and the GNU Mes RISC-V bootstrap project with Ekaitz Zarraga, Andrius Štikonas, and Jan Nieuwenhuizen. The bootstrap is now working from stage0 to tcc-boot0.
TinyCC compiles the RISC-V target, but still has some issues to resolve. The next steps include compiling the GNU C library, various versions of GCC, and packages beyond.
GNU Mes 0.25.1 was released with RISC-V support and a boostrappable-tcc branch. Both are available in Guix, though the RISC-V bootstrap is not yet enabled by default.
Outreach and User Support
Guix-HPC is in part about “spreading the word” about our approach to reproducible software environments and how it can help further the goals of reproducible research and high-performance computing development. This section summarizes talks and training sessions given this year.
Talks
Since last year, we gave the following talks at the following venues:
- Making reproducible and publishable large-scale HPC experiments, HPC & Big Data track, FOSDEM, Feb. 2024 (Philippe Swartvagher)
- Toward practical transparent, verifiable and long-term reproducible research using Guix, Institut Pasteur, Dec. 2023 (Simon Tournier)
- Reproducible software deployment in scientific computing, Event of the Max Planck Society, Sept. 2023 (Ricardo Wurmus)
- Guix: Funktionale Paketverwaltung zur wirklichen Reproduzierbarkeit, Second IT4Science Days, Meeting of the Helmholtz Association and the Max Planck Society, Sept. 2023 (Ricardo Wurmus)
- Building a Secure Software Supply Chain with GNU Guix, Programming Conference, March 2023 (Ludovic Courtès)
- Functional programming paradigm applied to package management: toward reproducible computational environment, IRILL, Feb. 2023 (Simon Tournier)
- Guix, toward practical transparent, verifiable and long-term reproducible research, Open Research Tools and Technology track, FOSDEM, Feb. 2023 (Simon Tournier)
- Vers une étude expérimentale reproductible avec GNU Guix, Rencontres sur les logiciels libres de recherche, Université de Strasbourg, Feb. 2023 (Marek Felšöci)
- Reproducibility and performance: why choose?, HPC & Big Data track, FOSDEM, Feb. 2023 (Ludovic Courtès)
To this list we should add 11 talks given for the First Workshop on Reproducible Software Environments for Research and High-Performance Computing, held in November 2023, for which videos are now on-line.
Events
As in previous years, Pjotr Prins and Manolis Ragkousis spearheaded the organization of the “Declarative and minimalistic computing” track at FOSDEM 2023, which was home to several Guix talks, along with the satellite Guix Days where 50 Guix contributors gathered.
This year, we held a second on-line reproducible research hackathon on reproducible research issues. This hackathon was a collaborative effort to leverage Guix to achieve reproducible software deployment for articles contributed to the online journal ReScience C. As outlined in our write-up on the experience, this served as an excellent opportunity to put into practice our guide to reproducible research papers, and it helped us identify open issues for long-term and archivable reproducibility.

This year we organized the First Workshop on Reproducible Software Environments for Research and High-Performance Computing, which took place in Montpellier, France, in November 2023. Coming from France primarily but also from Czechia, Germany, the Netherlands, Slovakia, Spain, and the United Kingdom to name a few, 120 people—scientists, high-performance computing (HPC) practitioners, system administrators, and enthusiasts alike—came to listen to the talks, attend the tutorials, and talk to one another.
Our ambition was to gather people from diverse backgrounds with a shared interest in improving their research workflows and development practices. The 11 talks and 8 tutorials, along with the hallway discussions and group dinner, have allowed us to share skills and experience. Videos of the talks edited by the video team at Institut Agro, our host, are available on the event’s web site.
Many thanks to our publicly-funded academic sponsors who made this event possible: ISDM, our primary sponsor for this event, Institut Agro for hosting the workshop in such a beautiful place, and EuroCC² and Inria Academy for their financial and logistical support. We look forward to organizing a second edition!
Training Sessions
For the French HPC Guix community, we continued the monthly on-line event called Café Guix, originally started in October 2021. Each month, a user or developer informally presents a Guix feature or workflow and answers questions. These sessions are now recorded and are available on the web page, gathering up to 70 people. This is continuing in 2024.
Pierre-Antoine Bouttier and Ludovic Courtès ran a 4-hour Guix training session as part of the User Tools for HPC (UST4HPC) event organized by CNRS (action nationale de formation, ANF) in June 2023. The session targeted an audience of HPC system administrators with no prior experience with Guix. Material (in French) is available on-line.
Marek Felšöci and Ludovic Courtès ran a 4-hour tutorial as part of the Compas HPC conference, in June 2023. The tutorial showed how to devise reproducible research workflows combining the literal programming facilities of Org-Mode with Guix. Supporting material is available on-line.
On September 27, Ricardo Wurmus hosted a 3-hour tutorial on the use of Guix for reproducible science as a session at the second IT4Science Days, a joint meeting of representatives of the Helmholtz Association of German Research Centres and the Max Planck Society. The workshop was attended by system administrators and scientists hailing from research institutes all over Germany.
The workshop on reproducible software environments that took place in Montpellier, France, in November 2023 was home to 8 tutorials, half of which about Guix. Each Guix tutorial had a different target audience: users-to-be (people with no prior experience with Guix), novice packagers, experienced packagers, and system administrators. Supporting material is available on the web page of the event.
A new MOOC on Reproducible Research practices has almost been completed. It will be stress-tested in February 2024 and open to the public on the platform FUN in spring. One of its three modules is about reproducible computational environments, introducing the various obstacles to reproducibility and presenting practical solutions. One of them is Guix, and in particular Guix containers defined by manifest files and frozen in time through channel files. Exporting such containers to Docker and Singularity is also discussed, because of the importance of these technologies in HPC.
Personnel
As part of Guix-HPC, participating institutions have dedicated work hours to the project, which we summarize here.
- Inria: 3.5 person-years (Ludovic Courtès and Romain Garbage; contributors to the Guix-HPC channel: Emmanuel Agullo, Julien Castelnau, Luca Cirrottola, Marek Felšöci, Marc Fuentes, Nathalie Furmento, Gilles Marait, Florent Pruvost, Philippe Swartvagher; system administrator in charge of Guix on the PlaFRIM and Grid’5000 clusters: Julien Lelaurain)
- University of Tennessee Health Science Center (UTHSC): 3+ person-years (Efraim Flashner, Bonface Munyoki, Fred Muriithi, Arun Isaac, Andrea Guarracino, Erik Garrison and Pjotr Prins)
- CNRS: 0.2 person-year (Konrad Hinsen)
- CNRS and Université Grenoble-Alpes (GRICAD): 0.2 person-year (Céline Acary-Robert, Pierre-Antoine Bouttier)
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC): 2 person-years (Ricardo Wurmus, Navid Afkhami, and Mădălin Ionel Patrașcu)
- Université Paris Cité: 0.75 person-year (Simon Tournier)
Guix itself is a collaborative effort, receiving code contributions from about 100 people every month, along with lots of crucial non-coding contributions: organizing events, writing documentation, giving tutorials, and more.
Perspectives
As the second decade dawns on the GNU Guix project, we shall take the opportunity to not only look back on past achievements, but evaluate our current position with respect to our goals and adjust our trajectory if necessary. Previous issues of the Activity Report had a common refrain: the importance of continuous efforts to connect the communities that meet at the intersection of Open Science, reproducible research, software development, system administration, and systems design. This issue is no different—the Guix-HPC effort remains committed to strengthening the ability of these communities to establish practices that further Open Science and make reproducible research workflows accessible.
The workshop on reproducible software environments in Montpellier may serve as an example of what this may look like in practice. The presenters in these sessions discussed issues of reproducible research and showcased the various roles Guix can assume in a diverse community of research practitioners: whether as the core of a platform for ad-hoc research environments; as the nexus that binds medical data, the tools of interpretation, and the scientific publication; or as the workhorse for reliably deploying entire HPC sites. As a project whose development priorizes increasing user autonomy, Guix has clearly found its niche among enthusiastic Open Science practitioners in a wide range of scientific fields.
While these activities are certainly encouraging, we need to acknowledge the fact that this level of engagement is not representative of the impact Guix has had on the wider scientific community. Challenges remain in bringing all the benefits and guarantees that Guix provides to where researchers actually do their computing, to the systems that system administrators get to build and maintain, and to the existing platforms and networks that represent the landscape in which computer-aided research takes place.
On the technical side, this could mean to contribute extensions to existing workflow systems like Snakemake or Nextflow; to develop tools and implement adapters for deploying Guix containers and virtual machine images to platforms like OpenStack; or to bridge gaps to support users of commercial third-party cloud computing platforms whose moats remain difficult to cross without leaving user autonomy behind.
These technical goals are, of course, informed by the needs of members of the reproducible research community who are currently represented in the Guix-HPC efforts. In the coming year, we want to continue to reach out to the wider community by organizing training sessions and workshops, and to gain better insight into how we can improve Guix to serve their needs. It is our mission to put the tools we build in the hands of practitioners at large—and to shape these tools together. Let’s talk—we’d love to hear from you!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).





