Guix-Jupyter 0.3.0 released!
We are pleased to announce Guix-Jupyter 0.3.0, a long-overdue release of our Guix-powered Jupyter kernel for self-contained and reproducible notebooks.

This comes after a three-year hiatus (more on that below) and was largely motivated by recent discussions on notebooks that revealed that yes, support for reproducible deployment in Jupyter Notebook is still much needed, at a time where the vast majority of published notebooks are not reproducible! It’s also the first release made from its new home at Codeberg.
Getting it
You can obtain it straight from Guix and spawn Jupyter Notebook with:
guix shell jupyter guix-jupyter -- jupyter notebookAlternatively, you can get the source from Git:
git clone https://codeberg.org/guix-science/guix-jupyter
cd guix-jupyter
git checkout v0.3.0 # or dc8cc183a573c63a07863d70ecff1fbf64e1dfc3
git tag -v v0.3.0Running guix shell --pure (with no arguments) gives you an interactive
development environment.
What’s new?
True to its mission of bringing reproducible deployment to the execution environment of Jupyter notebooks, this release adds new features and improvements.
Previous versions included the ;;guix pin “magic”, which lets you
pin (or lock) the guix channel—the main
channel
of Guix, with its 30k packages or so. However it lacked a way to
specify additional channels to get access to more packages.
This is now solved with the new ;;guix channel magic:
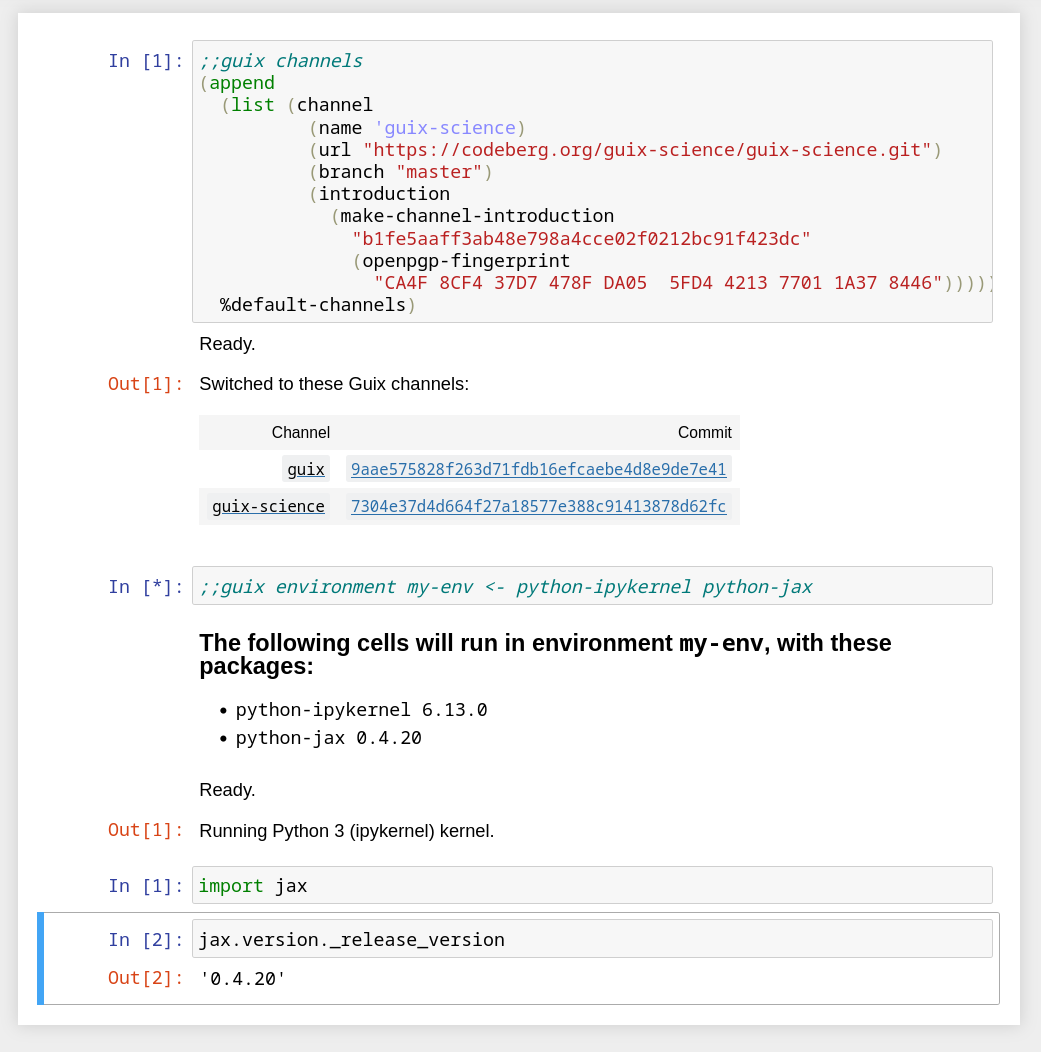
In the example above, we fetch the latest version of the
Guix-Science channel
together with the latest version of the Guix channel, following the
instructions for adding new
channels. By passing a
channel spec that includes commit references, we can pin both
channels and any channels they depend
on.
Other noteworthy improvements are in the built-in kernel for Guile programming. One of them is, at long last, support for completion:
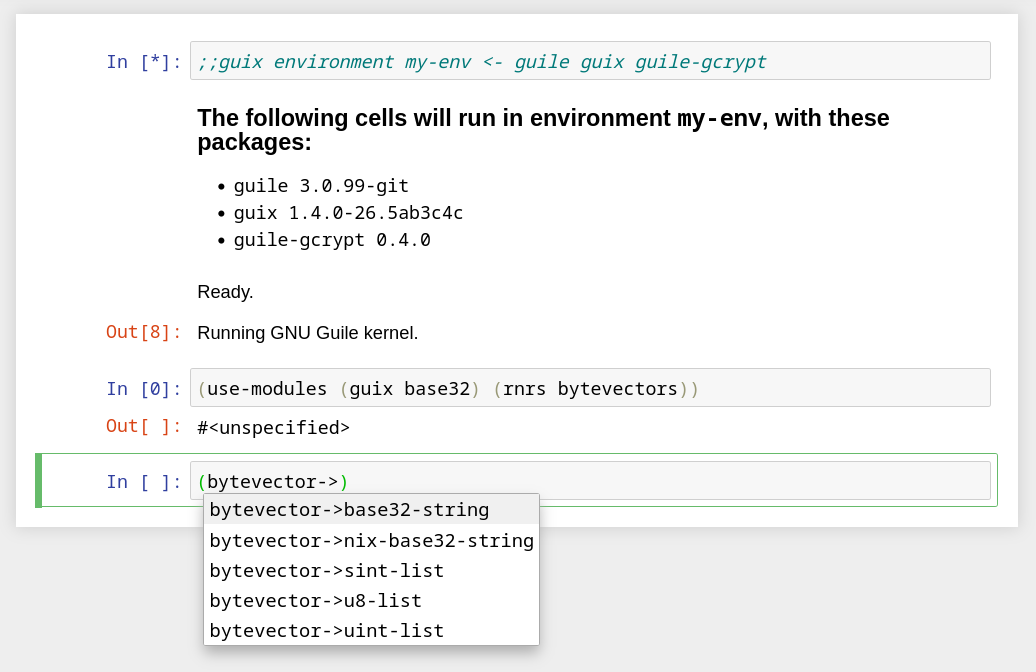
Check out the
NEWS
file for more info.
Make it yours!
The project has been dormant for the past couple of years. This is largely because the previous version “did the job”, but also because yours, truly, works primarily with HPC practitioners who are more into Org-Mode and/or fiddling at a terminal connected to their favorite supercomputer.
It remains that Jupyter is a popular and pleasant tool for reproducible research workflows, but one that desperately lacks a solution to deploy the execution environment of notebooks in a reproducible fashion. Guix-Jupyter is perhaps the only one of this kind. It is more than a mere proof of concept at this point but it can certainly benefit from user feedback.
With the move from Inria’s GitLab instance to
Codeberg, reporting
issues and submitting merge requests is finally made easier. Do not shy
away from contributing! If you’d like to get involved, we can also get
in touch on the guix-science mailing
list or on the
usual Guix communication channels.
Until then, happy hacking with Jupyter!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).





