Guix-HPC Activity Report, 2024

This document is also available as PDF (printable booklet)
Guix-HPC is a collaborative effort to bring reproducible software deployment to scientific workflows and high-performance computing (HPC). Guix-HPC builds upon the GNU Guix software deployment tools and aims to make them useful for HPC practitioners and scientists concerned with dependency graph control and customization and, uniquely, reproducible research.
Guix-HPC started as a joint software development project involving three research institutes: Inria, the Max Delbrück Center for Molecular Medicine (MDC), and the Utrecht Bioinformatics Center (UBC). Guix for HPC and reproducible research has since received contributions from many individuals and organizations, including CNRS, Université Paris Cité, the University of Tennessee Health Science Center (UTHSC), Cornell University, and AMD. HPC remains a conservative domain but over the years, we have reached out to many organizations and people who share our goal of improving upon the status quo when it comes to software deployment.
This report—our seventh report!—highlights key achievements of Guix-HPC between our previous report a year ago and today, February 2025. This year was marked by exciting developments for HPC and reproducible workflows. Significant advances were made in integrating Guix into the complex software landscape of HPC, taking the roles of software manager, workflow execution engine, backend for generating container images, or provider for the complete operating system layer. Support for reproducing computations from the past was also much improved. And, as usual, we have been using Guix for research, and teaching other researchers how to get started.
Outline
Guix-HPC aims to tackle the following high-level objectives:
- Reproducible scientific workflows. Improve the GNU Guix tool set to better support reproducible scientific workflows and to simplify sharing and publication of software environments.
- Cluster usage. Streamlining Guix deployment on HPC clusters, and providing interoperability with clusters not running Guix.
- Outreach & user support. Reaching out to the HPC and scientific research communities and organizing training sessions.
The following sections detail work that has been carried out in each of these areas.
Reproducible Scientific Workflows
Supporting reproducible research workflows is a major goal for Guix-HPC. This section looks at progress made on packaging and tooling.
Packages
With now more than 57,000 packages in Guix and related channels for scientific packages, the least we can say is that the package collection has grown significantly. Here are some of the highlights of the many additions that were made.
A critical element for HPC is libraries implementing the Message Passing Interface (MPI). Open MPI version 5 was added this year, together with upgrades of key packages: slurm, libfabric, ucx, and more. Noticeably, libfabric gained support for the HPE/Cray Slingshot interconnect. HPE released its supporting library, libcxi, in November 2023, and Guix was the first distribution to provide it. Slingshot support is particularly important for today’s HPC workloads: Slingshot is the interconnect found on key supercomputers, from Adastra (Tier-1 supercomputer in France), to LUMI (Tier-0 supercomputer operated by EuroHPC in Finland), and Frontier (currently the first supercomputer in Top500, hosted at ORNL in the United States).
We verified that our MPI packages achieve performance portability by
shipping MPI benchmarks bundled with guix pack to Adastra and running
them without further ado, and do not suffer performance loss compared to
the vendor-provided opaque MPI binaries. See the section Cluster Usage and
Deployment for concrete results obtained on major supercomputers
with different types of interconnects.
In the Guix-HPC channel,
the ROCm/HIP software stack for AMD GPUs was upgraded to version 6.2.
AMD engineers also contributed more packages to the collection,
including rocprof-register and hiprand (helper HIP/ROCm libraries)
and AOCL optimized numerical libraries for AMD processors (aocl-utils,
aocl-lapack, aocl-scalapack).
An increasingly important tool for computational sciences is the Julia programming language. In the main Guix channel, more than 300 packages of Julia libraries are now available, making it more readily usable.
Much scientific software packaging activity this year happened in the Guix-Science channel. First, we migrated the Guix-Science umbrella (which includes several channels in addition to Guix-Science) to Codeberg, a modern free software forge. We developed the missing parts to hook repositories at Codeberg into our continuous integration and continuous delivery (CI/CD) server at https://guix.bordeaux.inria.fr.
The Guix-Science channel received contributions for packages in new scientific domains, including physics, neuroscience, and electronics design. We are eager to see the list of scientific domains covered by Guix-Science grow.
Last, a new version of Guix-Jupyter, the Jupyter kernel that brings reproducible software deployment to notebooks, was released in November. The new version allows users to pin multiple channels in their notebook and improves its built-in kernel for Guile programming.
Concise Common Workflow Language
The Concise Common Workflow Language
(ccwl) is a concise syntax
to express Common Workflow Language (CWL)
workflows. It is implemented as an EDSL (Embedded Domain Specific
Language) in Guile Scheme. Unlike workflow languages such as the Guix
Workflow Language (GWL), ccwl is agnostic to deployment. It does not
use Guix internally to deploy applications. It merely picks up
applications from PATH and thus interoperates well with Guix and any
other package managers of the user's choice. ccwl also compiles to
CWL and thus reuses all tooling built to run CWL workflows. Workflows
written in ccwl may be freely reused by CWL users without impediment,
thus ensuring smooth collaboration between ccwl and CWL users.
ccwl 0.4.0 was released in January
2025. ccwl 0.4.0 provides a new
js-expression contruct to express CWL ExpressionTool class
workflows, and comes with more practical examples in the Cookbook
section of the manual.
ravanan
ravanan is a new Common Workflow Language (CWL) implementation that is powered by GNU Guix and provides strong reproducibility guarantees. ravanan runs CWL workflow descriptions as jobs on a HPC batch system. ravanan uses GNU Guix to manage all packages used by steps in the workflow. In addition, it takes inspiration from GNU Guix and uses a content-addressed store to provide strong caching of intermediate results. Thanks to these, ravanan never runs the same computation twice and its cache never goes stale.
Other salient features of ravanan include:
- One-to-one correspondence between steps in the CWL workflow and jobs on the HPC batch system;
- Clear logging for every job run;
- Jobs never write directly to the shared network filesystem on HPC, which is essential for good performance;
- Jobs never write to
/tmp.
Lack of reproducibility and increasingly complex software are a growing concern in scientific workflows. Many of these challenges have been effectively addressed by GNU Guix. ravanan takes this to the next level by seamlessly integrating Guix with a well-established standards-based workflow language like CWL.
ravanan had its first release—version 0.1.0—in January 2025.
Ensuring Source Code Availability
In June 2024, we published a new research paper entitled Source
Code Archiving to the Rescue of Reproducible
Deployment for the ACM
Conference on Reproducibility and
Replicability. The paper presents
work that has been done since we started connecting Guix with the
Software Heritage (SWH)
archive
five years ago. This is the first paper describing the design and
implementation of
Disarchive, our ‘tarball’
metadata extraction and recovery tool that, combined with Software
Heritage, allows Guix to automatically recover tar.gz and similar
source code files, even when they vanished from their original hosting
site.
Leading to this publication, we made significant strides in collaboration with the SWH development team to improve Guix’s ability to recover source code. In particular, Guix can now look up any version control checkout by content hash using the new SWH “ExtID” interface. Source code distributed with the Subversion version control tool was previously archived at SWH but unrecoverable by Guix; these changes fill the gap.
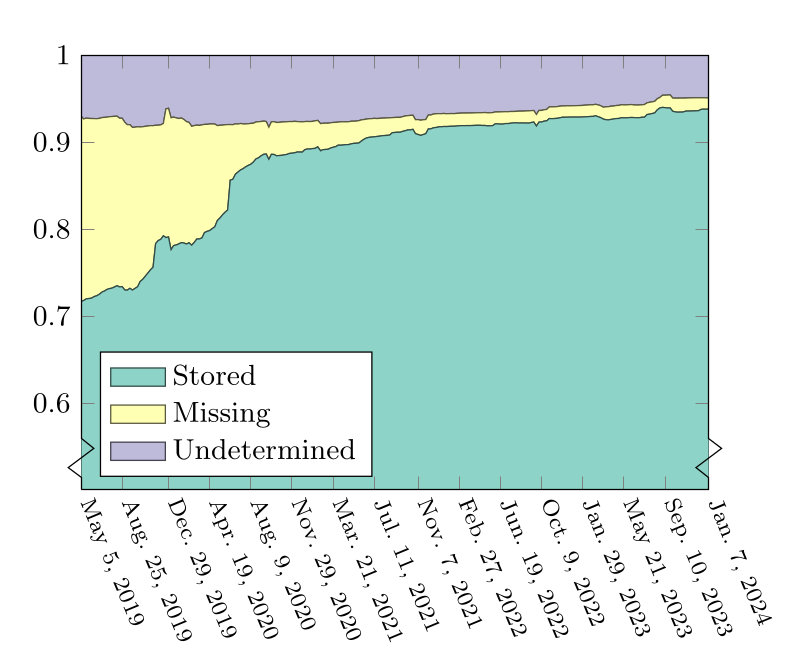
Timothy Sample’s Preservation of Guix reports further allowed us to monitor archival coverage and to identify discrepancies in Guix, Disarchive, and/or SWH. What these reports and the research article show is that, by addressing a number of issues, Guix as of January 2024 had 94% of its package source code archived.
Reproducible Research in Practice
In this section, we look at the variety of ways Guix is used to support reproducible research work.
Deploying Software from the Past
Ensuring source code availability, as discussed above, is just the first step to supporting reproducible deployment. Guix 1.0.0 was released in 2019 and our goal is to allow users to travel as far back as 1.0.0 and redeploy (and potentially rebuild) software from there, as in this example:
$ guix time-machine -q --commit=v1.0.0 -- \
environment --ad-hoc python2 -- python
> guile: warning: failed to install locale
Python 2.7.15 (default, Jan 1 1970, 00:00:01)
[GCC 5.5.0] on linux2
Type "help", "copyright", "credits" or "license" for more information.
>>>(The command above uses guix environment, the predecessor of guix shell,
which didn’t exist back then.)
It’s only 5 years ago but it’s pretty much remote history on the scale
of software evolution.
In the best case, the time-machine command above succeeds quickly,
thanks to the availability of substitutes (pre-built binaries) for that
2019 software stack. In the worst case, users have to rebuild some or
all of that software because substitutes are unavailable.
Unfortunately, while relying on isolated build
environments
prevents a number non-reproducibility issues by construction, a couple
of issues may still crop up. Chief among them are time traps:
software build processes that fail after a certain date.

Time traps are rare but not uncommon; historically they affected a few key packages: OpenSSL, GnuTLS, OpenJDK, Python, and more. The problem is that, because of them, attempts to rebuild today a software stack that contains one of these packages would fail.
We developed a solution to sidestep time traps and similar build issues. The new virtual build machine service for Guix System provides even more isolation for builds, and control over aspects that the Guix build daemon alone cannot afford. In particular, the service lets users choose the initial date of the virtual build machine and the CPU model it emulates. Once instantiated, Guix transparently offload builds to the virtual build machine, thereby allowing users to rebuild software packages from the past. This is an important improvement for reproducible research, though probably not the end of the road.
Supporting Artifact Evaluation at SuperComputing 2024
The Reproducibility
Initiative
of the International Conference for High Performance Computing, Networking,
Storage, and Analysis 2024 (SC24) proposed
the use of Guix as one the three official options for the
Artifact Evaluation (AE) process.
Because using the experimental
platform Chameleon Cloud is encouraged for the SC24 AE
process,
we have also designed and provided a Chameleon Cloud Guix-ready image, named
Guix-Ubuntu22.04-CUDA12.3-20240617.
We refer the blog article for an overview and to the the tutorial for more information.
DIAMOND and Guix
DIADEM is a French national exploratory program aiming at accelerating the discovery of new materials, launched at the end of 2023 with a budget of 85M€ for 6 years. Its numerical backbone, DIAMOND, is meant to provide a robust infrastructure on which all databases and simulation tools (for computation and visualisation) can rely. For the latter, a strong need was expressed for easily controllable and reproducible software environments. For the end user, the chosen solution is containerization, with Apptainer being the preferred candidate due to its HPC-friendly features and its already decent availability on the French HPC grid. For the time being, Guix acts primarily as a backend in the project, in order to build such containers via the guix pack command, de facto ensuring long-term reproducibility. At the time of writing this report, dozens of simulation tools covering all scales of materials simulation (see the code cloud below) are already available on the dedicated public channel. In addition, a “package to container image” conversion pipeline has also been set up, taking full advantage of GitLab-CI to automate the build, update, and deploy processes.
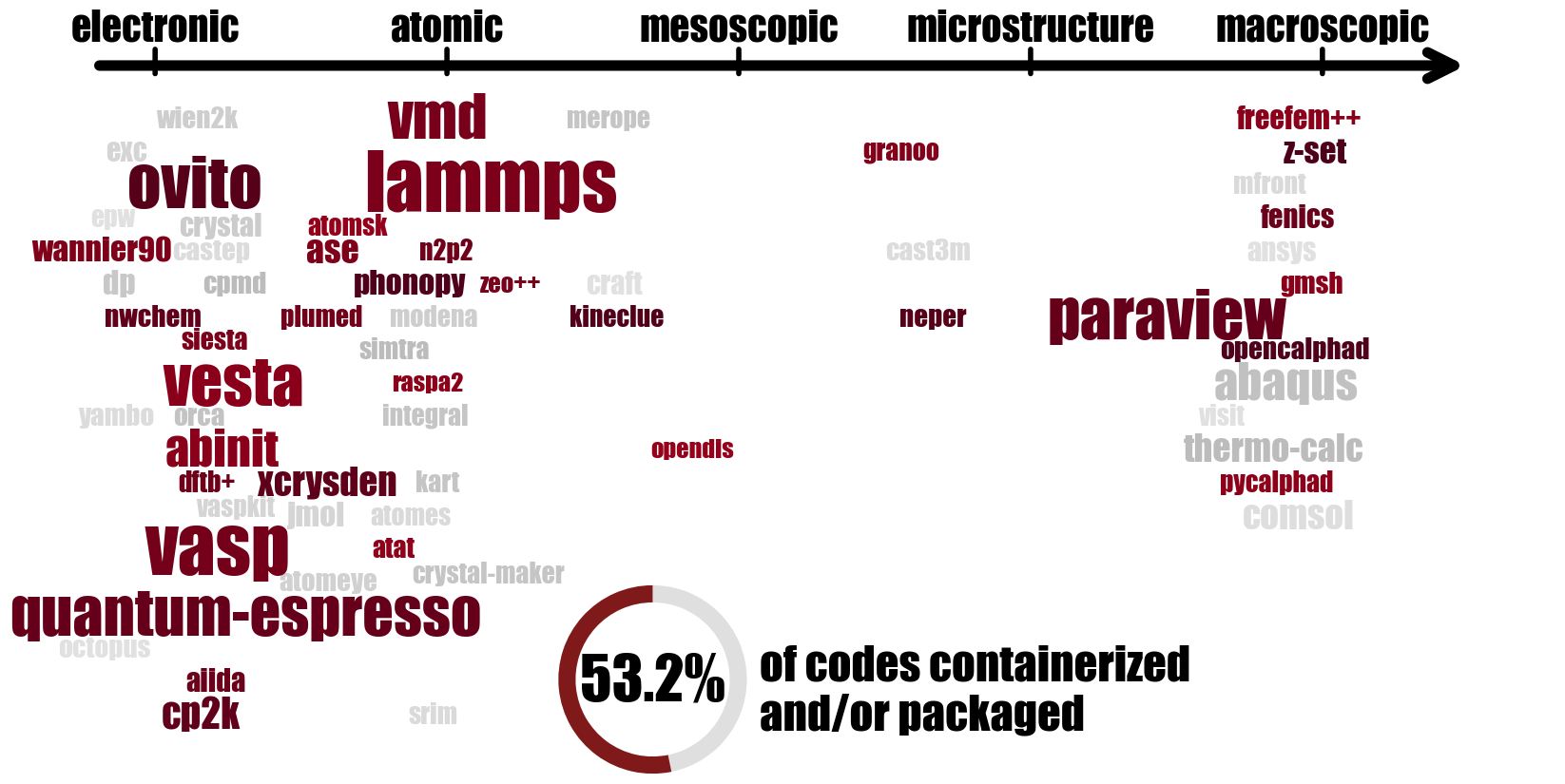
As well as filling out the list of available packages even more, there are also plans to give them better exposure, to escape from their current backend status, through extended documentation and upstream inclusion into other popular channels. The underlying hope is to follow the ever-growing Guix trend in the French HPC community (see for instance the NumPEx project or the MesoNET initiative, both using Guix) with a ready-to-use, robust set of popular tools for the materials simulation community. Another ongoing work in the DIAMOND project concerns providing ready-to-use workflows (chains of calculations) involving multiple simulation tools using AiiDA, a popular workflow manager in the materials science community.
Digital Electronics Design
A new research approach to digital electronics design of FPGAs has
recently arisen. This approach, strongly based on Guix, features
advanced packaging, versioning and dependency management capabilities
for gateware products in the form of VHDL design units. This departs
from traditional methods in this domain, based on Tcl or Python
scripting as a glue to the several existing building blocks. Using Guix,
it becomes simple to follow a declarative paradigm to handle firmware
dependencies, in parallel to creating reproducible software
environments, which guarantees traceability during the full design
cycle. Thanks to last year's updates to the main Guix repository, but also to
new packages in the Guix-Science channel and the
Electronics channel,
it is now possible to elaborate and simulate (GHDL and NVC compilers) designs in
VHDL-2008, making use of unit testing frameworks (vunit) and modern
verification libraries (osvvm), in addition to performing cosimulation
(cocotb). It is also feasible to synthesize and to implement formal
verification of complex logic (yosys) in native VHDL (ghdl-yosys-plugin).
In parallel to all previous additions, Guix's performance, flexibility and stability as the base of a continuous integration framework are currently being investigated in the context of FPGA verification. This way, sophisticated pipelines using the Guix Workflow Language (GWL) are being deployed to remote build facilities on public Git forges. Based on native Guix System images (for SourceHut), updated daily, or on user-defined custom Docker containers (for GitLab), several testing pipelines are being deployed to verify VHDL designs. An introduction to current developments was presented by Cayetano Santos during the latest CNRS/CEA system administration and software engineering conference (15ème Journées Informatiques IN2P3/IRFU).
Reproducible Multiphysics Simulation and Workflow Runs on HPC Systems
As part of efforts to enable reproducible workflows across diverse HPC environments, a team at the Helmholtz Centre for Environmental Research – UFZ implemented an AiiDA-based workflow to create portable, optimized, and reproducible container images of the OpenGeoSys software suite which enable consistent multi-physics simulations and complex workflow runs on various HPC platforms.
Using GNU Guix, the software stack was generated on multiple machines from a source bootstrap, ensuring bit-by-bit reproducibility of the resulting Apptainer images. These images were then deployed to three HPC systems: JUWELS at the Jülich Supercomputing Centre (Tier-0), BARNARD at TUD Dresden University of Technology (Tier-2), and EVE at the Helmholtz Centre for Environmental Research – UFZ. The containers were evaluated on all environments through MPI interconnect performance checks, simple MPI-based simulations (3 cores), multi-node MPI-based simulations (96 cores), and a complex Snakemake-based workflow from the AREHS project running on all cores of a single node. All outputs, including simulation results and plots generated by the Snakemake workflow, are bit-by-bit identical reproduced on all three HPC platforms.
In summary, GNU Guix enabled the reliable reproduction of complex multiphysics simulation results across multiple HPC systems using only the original input data and the commit hashes of Guix and related package channels to generate the required software environment.
Exploring the Impact of Hardware Variability
In a paper presented at ACM REP'24 that received the “Best Paper Award”, we studied the effect of hardware variability on results produced by the FSL FLIRT application, a widely-used software component in neuroimaging data analyses.
While software containerization solutions such as Docker and Singularity have been deployed to mask the effects of software-induced variability, variations in hardware architectures still impact neuroimaging results in an unclear way. Using the Grid’5000 infrastructure, we studied the effect of nine different CPU models using two software packaging systems (Docker and Guix), and we compared the resulting hardware variability to numerical variability measured with random rounding. Results showed that hardware, software, and numerical variability lead to perturbations of similar magnitudes. For the hardware variability, differences in results were due to differences in micro-architectures, specifically the presence or absence of AVX-2 support. The effect of hardware perturbations on linear registration remained moderate, but might impact downstream analyses when linear registration is used as initialization step for other operations.
In the course of this study, we also wrote FSL Guix modules to compile the FSL application and all its dependencies in a reproducible manner. FSL Guix modules are available on a public Git repository.
Cluster Usage and Deployment
The sections below highlight the experience of cluster administration teams and report on tooling developed around Guix for users and administrators on HPC clusters.
MPI Performance Portability
MPI performance portability — achieving optimal MPI performance on all supported hardware — is a long-standing goal of the Guix-HPC project. Progress has been made towards the achievement of this goal.
Support for the Cray/HPE Slingshot interconnect in Open MPI through libfabric/libcxi was added as soon as the code was open-sourced.
The libfabric package definition has been improved to support a broader variety of network interconnects: it now supports Intel Omni-Path through PSM2, Infiniband, iWarp and RoCE through the Linux Verbs API. The MPICH implementation of MPI can now use these interconnect drivers.
CUDA support has been improved: Open MPI has now GPUDirect support and
the nccl library is now available.
Tests have been carried out on French Tier-1 supercomputers (Adastra and Jean-Zay) to ensure proper performance was achieved by the MPI software stack. Specifically, NVIDIA GPUDirect/NCCL and ROCm RCCL support has been tested using the OSU Micro Benchmarks suite.
Tier-0 and Tier-1 Supercomputers
Reproducible software deployment using Guix on French Tier-1 and EuroHPC Tier-0 machines is one of the goals of the Development and Integration work package of NumPEx, the French exascale HPC program, the main constraint being that Guix is not available on these machines.
Progress has been made towards this goal, particularly on TGCC's Irène
cluster, where Guix is now able to produce Docker container images
suitable for pcocc, TGCC's
container deployment tool.
Guix offer different ways of generating a software stack that can then be deployed on a supercomputer:
- Singularity/Apptainer image generation using
guix pack -f squashfs - Relocatable binary archive generation using
guix pack -RR - Docker image generation using
guix pack -f docker
Below is a summary of the techniques that have been validated on the different target machines, French national supercomputers (so-called “Tier-1”) and EuroHPC supercomputers (“Tier-0”):
- Jean-Zay (IDRIS): Singularity images and relocatable binary archives.
- Adastra (CINES): Relocatable binary archives (Singularity, which is also available, has not been tested but should work out-of-the box).
- Irène (TGCC): Docker images (currently validated for single
processes only), both on
x86_64andaarch64CPU architectures. - LUMI (EuroHPC): Singularity images (currently validated on a single node).
As an example of successful application of these techniques, AVBP, a physics simulation code that has been packaged in Guix, has been run on Adastra on up to 1,600 MPI processes (80 processes per node on 20 nodes) using a relocatable binary archive and on Jean-Zay (up to 256 MPI processes, on 256 nodes with a single process per node) using a Guix generated Singularity image.
A number of tutorials related to reproducible software deployment on Tier-1 and Tier-0 clusters have been published by the NumPEx Development and Integration work package.
Future work will be needed to validate multi-processes/multi-nodes on TGCC and EuroHPC machines.
Pangenome Genetics Research Cluster at UTHSC
At UTHSC, Memphis (USA), we are running a 16-node large-memory Octopus HPC cluster (438 real CPU cores) dedicated to pangenome and genetics research. In 2024, the storage doubled up to a 400 TB Lizardfs SSD fiber-optic connected distributed network storage. We aim to replace the unmaintained Lizardfs software with a more efficient Ceph distributed storage.
Notable about this HPC cluster is that it is administered by the users themselves. Thanks to Guix, we install, run and manage the cluster as researchers — and roll back in case of a mistake. UTHSC IT manages the infrastructure—i.e., physical placement, electricity, routers and firewalls — but beyond that there are no demands on IT. Thanks to out-of-band access, we can completely (re)install machines remotely. Octopus runs Guix on top of a minimal Debian install and we are experimenting with pure Guix virtual machines and nodes that can be run on demand. Almost all deployed software has been packaged in Guix and can be installed on the head-node by regular users on the cluster without root access. This same software is shared through NFS on the nodes. See the guix-bioinformatics channel for all deployment configuration.
Thanks to Collin Doering and the wider Guix community we now have Guix on a bare metal machine that acts as a build `farm' for Guix North America that serves the continent and also services our HPC. Collin's blog goes into more detail in his post Setup of a Simple Guix Build Farm and Substitute Server.
Supporting RISC-V
RISC-V represents avanced open source hardware and is making inroads with HPC, e.g. in Barcelona and with the new Barcelona Supercomputing Center Sargantana chip. EU investments in RISC-V are coming online. Other countries are also investing deeply into RISC-V including China.
Christopher Batten (Cornell) and Michael Taylor (University of Washington) are in charge of creating the NSF-funded RISC-V supercomputer with 2,000 cores per node and 16 nodes in a rack (NSF PPoSS grant 2118709), targeting Guix driven pangenomic workloads by Erik Garrison, Arun Isaac, Andrea Guarracino, and Pjotr Prins. The aim is to have a prototype running in 2025.
The supercomputer will incorporate Guix and the GNU Mes bootstrap, with input from Arun Isaac, Efraim Flashner and others.
NLNet funds RISC-V support for the Guix riscv64 target from Efraim Flashner and the GNU Mes RISC-V bootstrap project with Ekaitz Zarraga, Andrius Štikonas, and Jan Nieuwenhuizen. The RISC-V bootstrap is fully working and can be adopted by Linux distributions!
Outreach and User Support
Guix-HPC is in part about “spreading the word” about our approach to reproducible software environments and how it can help further the goals of reproducible research and high-performance computing development. This section summarizes talks and training sessions given this year.
Articles
The following refereed articles about Guix were published:
- Ludovic Courtès, Timothy Sample, Simon Tournier, and Stefano Zacchiroli, Source Code Archiving to the Rescue of Reproducible Deployment, 2nd ACM Conference on Reproducibility and Replicability (ACM REP), June 2024
- Gael Vila, Emmanuel Medernach, Ines Gonzalez Pepe, Axel Bonnet, Yohan Chatelain, Michael Sdika, Tristan Glatard, and Sorina Camarasu Pop The Impact of Hardware Variability on Applications Packaged with Docker and Guix: a Case Study in Neuroimaging, 2nd ACM Conference on Reproducibility and Replicability (ACM REP), June 2024
- Simon Tournier, (Re)Déploiement de conteneurs et machines virtuelles avec Guix Journées Réseaux de l'Enseignement et de la Recherche (JRES), Dec. 2024
Talks
Since last year, we gave the following talks at the following venues:
- RISC-V Bootstrapping in Guix and Live-Bootstrap, FOSDEM, February 2024 (Ekaitz Zárraga)
- Making reproducible and publishable large-scale HPC experiments, FOSDEM, February 2024 (Philippe Swartvagher)
- Reproducibility and Performance: Why Choose?, 52nd ORAP Forum, March 2024 (Ludovic Courtès)
- Dependable Software Deployment with Guix, International Post-Exascale Workshop (InPEx), June 2024 (Ludovic Courtès)
- Reproducibility and replicability of computer simulations, Keynote at ACM REP 2024, June 2024 (Konrad Hinsen)
- Source Code Archival To The Rescue Of Reproducible Deployment, at ACM REP 2024, June 2024 (Simon Tournier)
- Reproducibilité des résultats de recherche à l’aide de GNU/Guix, webcast, 15èmes Journées Informatiques IN2P3/IRFU, Sep. 2024 (Cayetano Santos)
- Continuous Integration & Continuous Delivery for HPC with Guix, Webinar of the CASTIEL 2 European Project, Sep. 2024
- Reconciling high-performance computing with the use of third-party libraries?, Journées Calcul et Données (JCAD), Nov. 2024 (Emmanuel Agullo)
- (Re)Déploiement de conteneurs et machines virtuelles avec Guix Journées Réseaux de l'Enseignement et de la Recherche (JRES), Dec. 2024 (Simon Tournier)
- Environnement logiciel reproductible, à quelle échéance ?, 2ème Journée d'Étude ARDoISE, Dec. 2024 (Simon Tournier)
- Reproducible Software Deployment with Guix in HPC, Seminar at CERFACS, Dec. 2024 (Ludovic Courtès)
- Déployer AVBP avec Guix, Seminar at CERFACS, Dec. 2024 (Romain Garbage)
- Reproducible Software Deployment with GNU Guix, Seminar of the ADAC working group on Portability, Sustainability, and Integration, Jan. 2025 (Ludovic Courtès)
- At the Barcelona Supercomputing Center (BSC), Arun Isaac and Pjotr Prins presented their work on Guix for HPC, including ravanan, an optimized and reproducible Common Workflow Language (CWL) runner.
- … and the talks of the November 2024 Guix-HPC Workshop
Events
As has become tradition, Pjotr Prins and Manolis Ragkousis spearheaded the organization of the “Declarative and minimalistic computing” track at FOSDEM 2024, which was home to several Guix talks, along with the satellite Guix Days where 50 Guix contributors gathered.
As a followup to the workshop that took place in November 2023, we organized a Mini Workshop on Guix in HPC. The event took place in Bordeaux, France, with remote attendance as well, right after JCAD, the French-speaking conference for HPC practitioners. About forty people joined on site and a dozen more on-line. The aim of the workshop was twofold. The fist half day was dedicated to the users point of view and how to manage calculus with Guix in various contexts. During the second half day, presentations and discussions focused on the use of Guix as a package manager in mesocentres. Speakers showed the use of Guix in HPC in a variety of disciplines—from material physics to linear algebra and system administration—by PhD candidates, research software engineers, and senior system administrators. In addition to these talks, a panel allowed HPC center engineers to share their experience setting up Guix on clusters and discuss pain points and issues that would need to be addressed for broader adoption.
Training Sessions
Café Guix
Café Guix is an informal monthly discussion about the Guix software environment manager. Students, researchers, system administrators, support staff from laboratories or computing centres — everyone is welcome to join in this one-hour monthly meeting to discuss questions they have about Guix and its use in the broadest sense. Each session has a specific level of difficulty, clearly indicated in the program. Some sessions are designed for beginners and others for more experienced users. For example, in 2024, entry-level topics included an introduction of Guix from a user point of view and the presentation of basic commands. We also covered more advanced topics such as the use of G-exp and the different stages in the life of a package.
MOOC “Reproducible Research II: Practices and tools for managing computations and data”
A first session of this new online course (MOOC) has been run from 16 May to 26 September 2024. One of its three modules is about reproducible computational environments. It includes a detailed introduction to Guix, with exercises, and it uses Guix to ensure the reproducibility of its main example, a workflow for detecting sunspots in a database of thousands of images of the sun. A revised second session is planned for April 2025.
MDC seminar in the series “Research Data Management: Monday seminars on Reproducibility”: “Software reproducibility with Guix”
This seminar session took place on 22 April 2024 at the MDC for an audience of bioinformatics researchers. In this session we looked at the problem of computational reproducibility, clarified the role (and the limitations) of containers in the realm of reproducibility, and introduced the declarative approach to reproducible and portable software environments using Guix.
Personnel
GNU Guix is a collaborative effort, receiving contributions from more than 90 people every month—a 50% increase compared to last year. As part of Guix-HPC, participating institutions have dedicated work hours to the project, which we summarize here.
- CNRS: 0.2 person-year (Konrad Hinsen)
- Inria: 3.5 person-years (Ludovic Courtès and Romain Garbage; contributors to the Guix-HPC, Guix-Science, and Guix channels: Emmanuel Agullo, Julien Castelnau, Luca Cirrottola, Marc Fuentes, Gilles Marait, Florent Pruvost, Philippe Swartvagher; system administrator in charge of Guix on the PlaFRIM and Grid’5000 clusters: Julien Lelaurain)
- University of Tennessee Health Science Center (UTHSC): 3+ person-years (Efraim Flashner, Collin Doering, Bonface Munyoki, Fred Muriithi, Arun Isaac, Andrea Guarracino, Erik Garrison and Pjotr Prins)
- GRICAD : CNRS 0.5 person-year (Benjamin Arrondeau and Pierre-Antoine Bouttier), UGA 0.1 person-year (Céline Acary-Robert)
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC): 2 person-years (Ricardo Wurmus, Navid Afkhami, and Mădălin Ionel Patrașcu)
- Université Paris Cité: 0.5 person-year (Simon Tournier)
Perspectives
Computational reproducibility is still widely regarded as too difficult to achieve. In 2024, we have continued to lower the barriers, by improving the Guix itself and by integrating it with other foundational tools of computational science: workflow managers and widely deployed container runtimes. Integration with another foundational tool, continuous integration on software forges, remains unsatisfactory so far, but is being worked on. We have also demonstrated that reproducibility is not in contradiction with striving for performance. Scientists from a growing number of domains are adopting Guix, as illustrated by the introduction of new packages in the Guix-Science channel, targeting domains such as physics, neuroscience, or electronics design.
So far, we have focused on addressing the needs of two groups of actors in HPC:
- HPC users—i.e., researchers and engineers who use HPC in research projects;
- administrators of HPC machines.
There are at least two further groups of actors relevant for HPC:
- developers of scientific software deployed on HPC systems;
- vendors of HPC machines and the associated systems software.
What Guix-HPC could do for these two groups is provide support and training on reproducibility issues. Time traps, for example, should be avoided (or removed) by the upstream projects, rather than worked around at the packaging and deployment stages. Prevention is better than cure. But most software developers today are unfamiliar with the concept of time traps.
A much more ambitious challenge is working with developers of language-specific packaging and distribution systems in view of enhanced integration with system-level packaging approaches such as Guix. The JavaScript ecosystem is perhaps the best-known example of an ecosystem whose packaging habits make it difficult to build software reproducibly from source code (see this ten-year-old rant for an example). The Julia and Rust ecosystems are taking a similar direction. With the growing popularity of Julia and Rust in scientific computing, new obstacles to reproducibility are therefore appearing on the horizon.
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).







